#4 Data Release 2 Weekly Briefing
I am currently generating a list of tasks, implemented as issues, that can be found on our git repository. Each task will be populated with a detailed description, a team member assigned, and any perquisites needed to complete the task. A workflow of these tasks is described in Fig. 1.
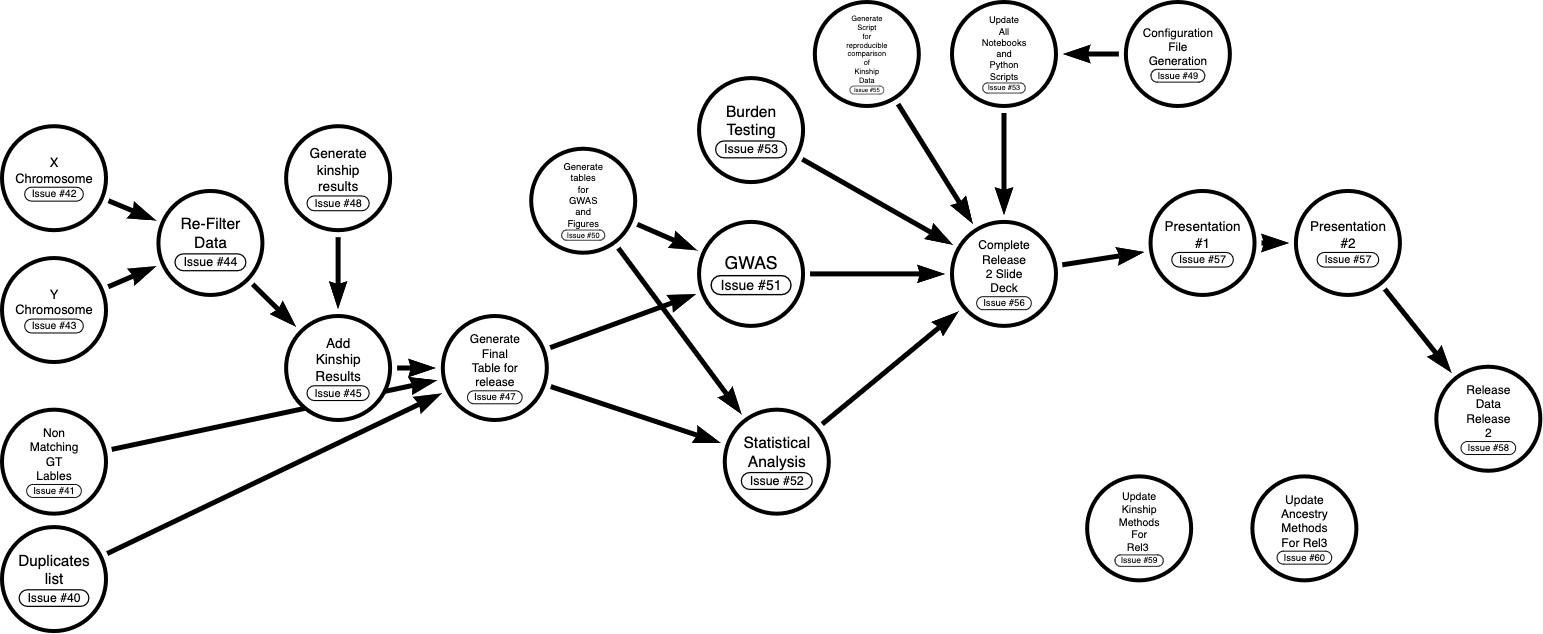
Three additional issues recently created are in relation to discussions we had in our last, (01/20/2023), group meeting. First, the question of whether mitochondrial DNA is included in our data came up. The answer to this question is no. It was also mentioned that the Boston team had used a minor allele frequency filtering cutoff of 5% to run kinship analysis. This is something our pipeline is lacking and may be worth adding for our own kinship analysis.
Lastly, a review of our genotype filtering may need to be conducted. We are currently filtering pseudoautosomal regions on the sex chromosomes as autosomes. Depending on how genome alignment and variant calling are conducted, this may need to be changed.
Discuss this update on our GitHub!