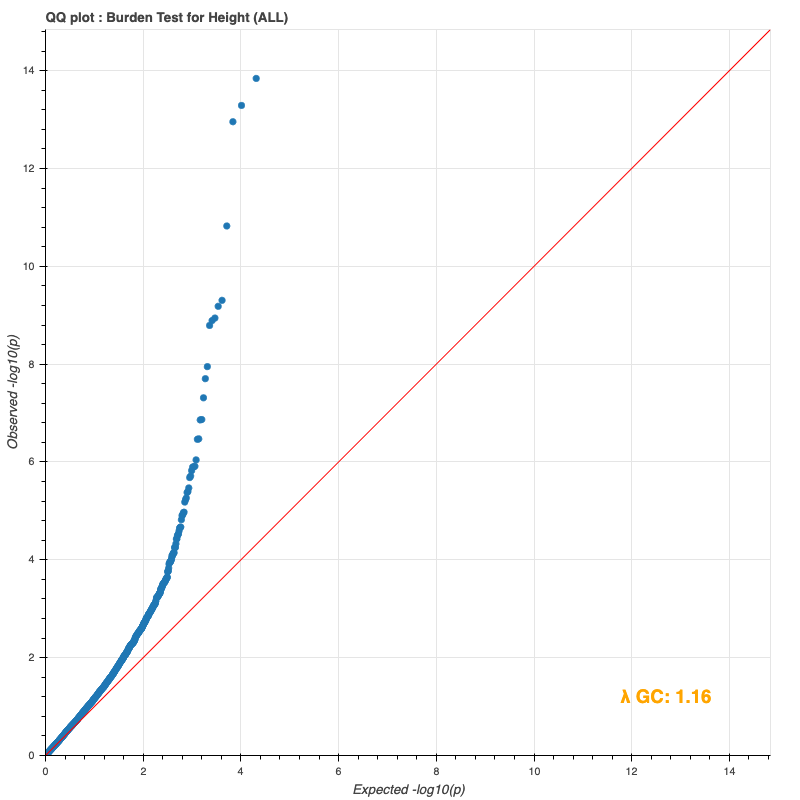
#27(b) Burden testing mini-update
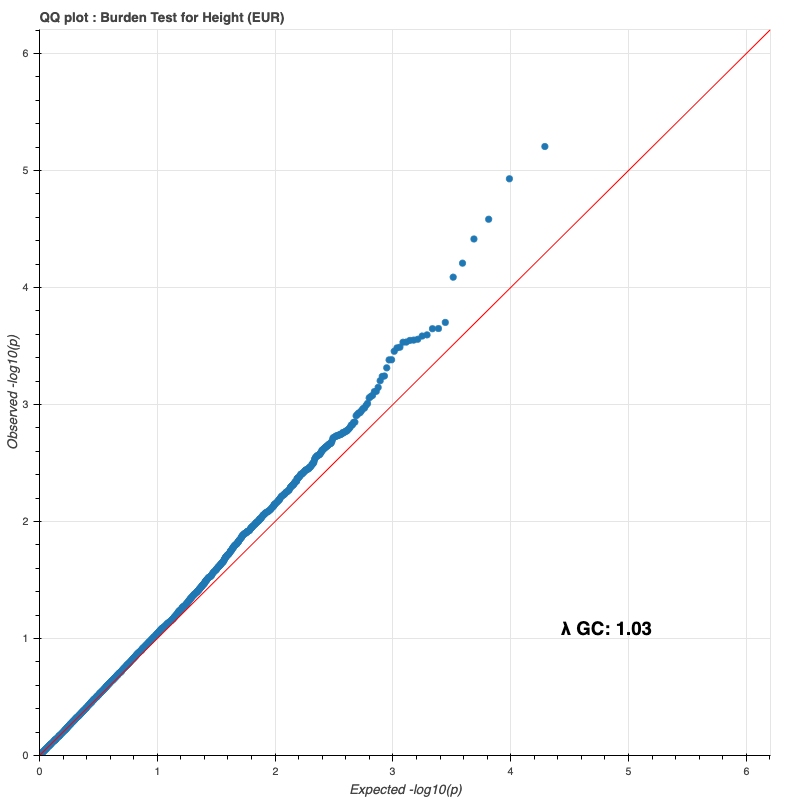
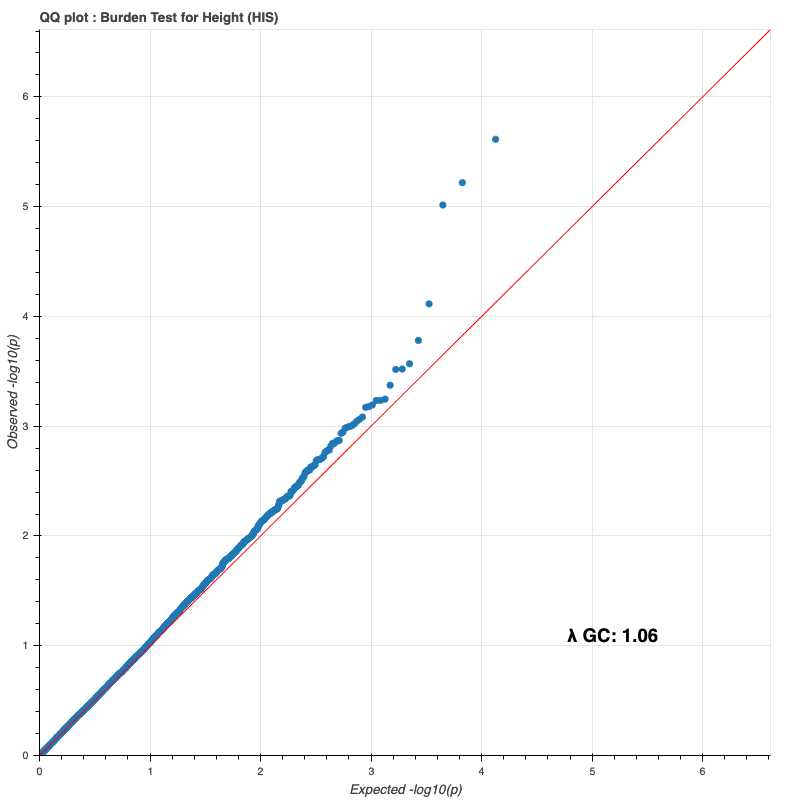
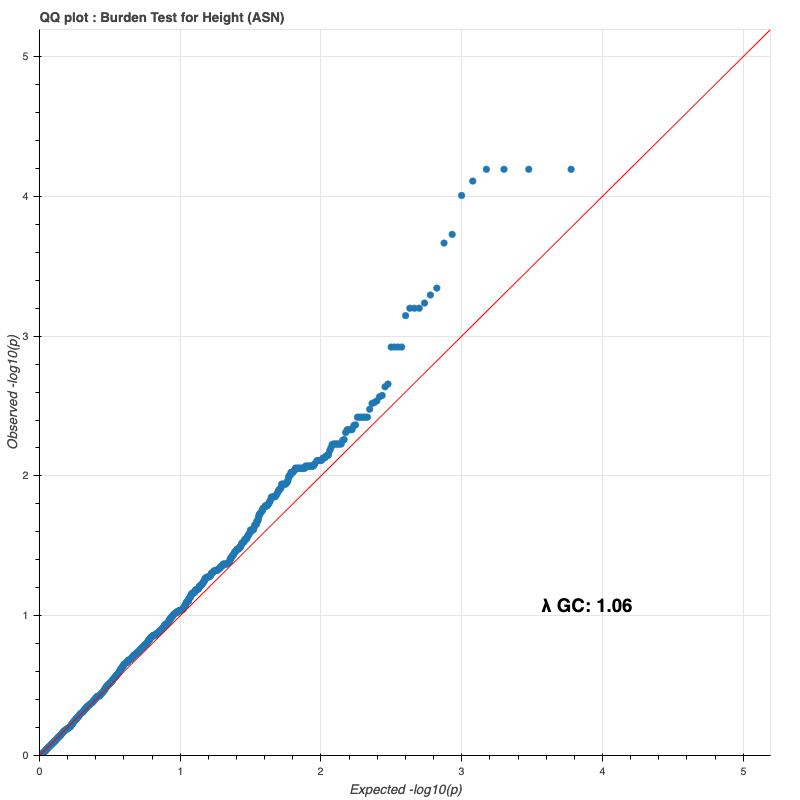
Since last time, I reran the burden test on the entire DR2 population and all subpopulations, using whole-population and per-ethnicity PCA scores, respectively. The results still do not match Bryan’s (I only compared against my European results against his, because Genebass didn’t provide a way to filter by subpopulation).
European subpopulation
| gene | n | sum_x | y_transpose_x | beta | standard_error | t_stat | p_value | bryan_gene_rank |
|---|---|---|---|---|---|---|---|---|
| “HBB” | 72152 | 6.10e+01 | 4.12e+03 | -1.52e+00 | 3.37e-01 | -4.52e+00 | 6.24e-06 | 3016 |
| “OR7A10” | 72152 | 3.00e+00 | 2.30e+02 | 6.65e+00 | 1.52e+00 | 4.38e+00 | 1.18e-05 | NA |
| “ZNF382” | 72152 | 1.20e+01 | 7.99e+02 | -3.19e+00 | 7.59e-01 | -4.21e+00 | 2.61e-05 | NA |
| “ACVR2B-AS1” | 72152 | 2.00e+00 | 1.55e+02 | 7.65e+00 | 1.86e+00 | 4.12e+00 | 3.85e-05 | NA |
| “LINC02193” | 72152 | 1.00e+00 | 8.05e+01 | 1.05e+01 | 2.63e+00 | 4.01e+00 | 6.19e-05 | NA |
| “XNDC1N-ZNF705EP-ALG1L9P” | 72152 | 4.74e+02 | 3.27e+04 | -4.77e-01 | 1.21e-01 | -3.94e+00 | 8.16e-05 | NA |
| “ZNF14” | 72152 | 9.60e+01 | 6.78e+03 | 9.99e-01 | 2.68e-01 | 3.72e+00 | 1.99e-04 | NA |
| “HBP1” | 72152 | 1.70e+01 | 1.14e+03 | -2.35e+00 | 6.37e-01 | -3.69e+00 | 2.24e-04 | 6534 |
| “GHSR” | 72152 | 1.00e+01 | 6.62e+02 | -3.07e+00 | 8.31e-01 | -3.69e+00 | 2.25e-04 | NA |
| “ZNF385C” | 72152 | 1.90e+01 | 1.36e+03 | 2.21e+00 | 6.03e-01 | 3.66e+00 | 2.54e-04 | 15163 |
Hispanic subpopulation
| gene | n | sum_x | y_transpose_x | beta | standard_error | t_stat | p_value |
|---|---|---|---|---|---|---|---|
| “LINC01607” | 5511 | 1.00e+00 | 5.60e+01 | -1.24e+01 | 2.63e+00 | -4.72e+00 | 2.43e-06 |
| “ATOSB” | 5511 | 2.00e+00 | 1.48e+02 | 8.42e+00 | 1.86e+00 | 4.53e+00 | 6.04e-06 |
| “LRRC49” | 5511 | 6.40e+01 | 4.24e+03 | -1.47e+00 | 3.32e-01 | -4.43e+00 | 9.65e-06 |
| “SCN5A” | 5511 | 4.00e+00 | 2.53e+02 | -5.20e+00 | 1.31e+00 | -3.96e+00 | 7.68e-05 |
| “AGPAT5” | 5511 | 1.00e+00 | 7.81e+01 | 9.92e+00 | 2.63e+00 | 3.77e+00 | 1.65e-04 |
| “CTBS” | 5511 | 2.00e+00 | 1.50e+02 | 6.78e+00 | 1.86e+00 | 3.65e+00 | 2.70e-04 |
| “TMEM198B” | 5511 | 1.00e+01 | 7.00e+02 | 3.01e+00 | 8.32e-01 | 3.62e+00 | 3.01e-04 |
| “LINC02210” | 5511 | 2.50e+01 | 1.73e+03 | 1.90e+00 | 5.27e-01 | 3.61e+00 | 3.04e-04 |
| “SNN” | 5511 | 1.06e+02 | 7.09e+03 | -9.23e-01 | 2.62e-01 | -3.53e+00 | 4.23e-04 |
| “BCL11A” | 5511 | 3.90e+01 | 2.71e+03 | 1.46e+00 | 4.22e-01 | 3.45e+00 | 5.67e-04 |
Asian subpopulation
| gene | n | sum_x | y_transpose_x | beta | standard_error | t_stat | p_value |
|---|---|---|---|---|---|---|---|
| “GRIK2” | 674 | 1.00e+00 | 7.69e+01 | 1.02e+01 | 2.53e+00 | .02e+00 | 6.40e-05 |
| “NOL4L-DT” | 674 | 1.00e+00 | 7.69e+01 | 1.02e+01 | 2.53e+00 | .02e+00 | 6.40e-05 |
| “NUDT13” | 674 | 1.00e+00 | 7.69e+01 | 1.02e+01 | 2.53e+00 | .02e+00 | 6.40e-05 |
| “S100A7A” | 674 | 1.00e+00 | 7.69e+01 | 1.02e+01 | 2.53e+00 | .02e+00 | 6.40e-05 |
| “RB1” | 674 | 4.00e+00 | 2.47e+02 | -5.03e+00 | 1.26e+00 | .98e+00 | 7.77e-05 |
| “TBATA” | 674 | 6.00e+00 | 3.84e+02 | -4.13e+00 | 1.06e+00 | .92e+00 | 9.85e-05 |
| “FZD5” | 674 | 4.00e+00 | 2.49e+02 | -4.76e+00 | 1.27e+00 | .76e+00 | 1.87e-04 |
| “MCMBP” | 674 | 2.00e+00 | 1.47e+02 | 6.67e+00 | 1.79e+00 | .72e+00 | 2.16e-04 |
| “MMP19” | 674 | 4.00e+00 | 2.83e+02 | 4.47e+00 | 1.27e+00 | .52e+00 | 4.53e-04 |
| “MUC5AC” | 674 | 2.00e+00 | 1.44e+02 | 6.30e+00 | 1.80e+00 | .49e+00 | 5.07e-04 |
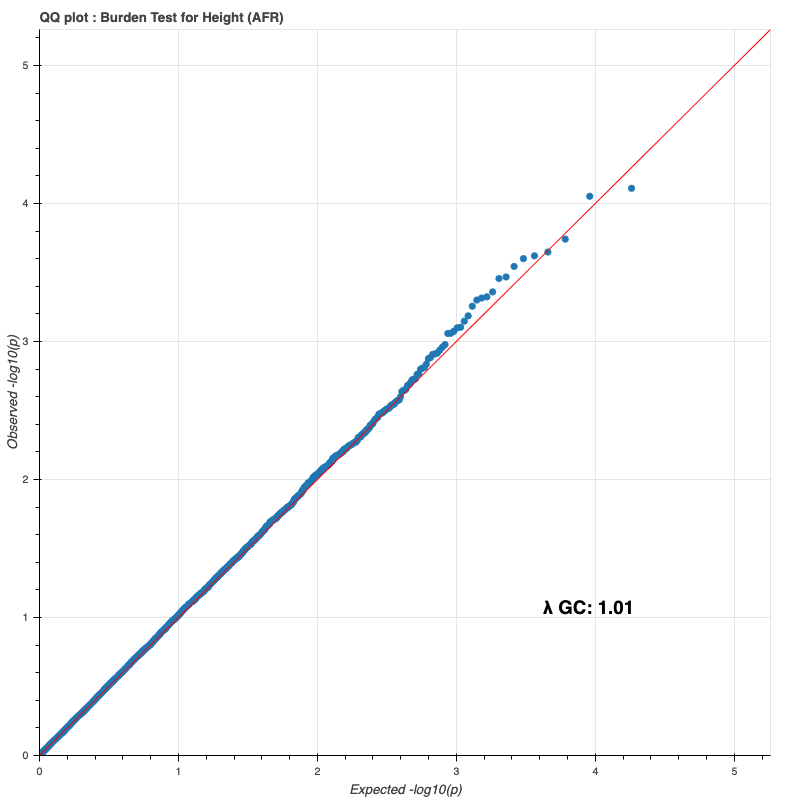
African subpopulation
| gene | n | sum_x | y_transpose_x | beta | standard_error | t_stat | p_value |
|---|---|---|---|---|---|---|---|
| “BRF1” | 24528 | 2.10e+01 | 1.51e+03 | 2.39e+00 | 6.04e-01 | 3.95e+00 | 7.76e-05 |
| “LINC01435” | 24528 | 6.20e+01 | 4.24e+03 | -1.38e+00 | 3.52e-01 | -3.92e+00 | 8.86e-05 |
| “SLCO4C1” | 24528 | 3.60e+01 | 2.57e+03 | 1.73e+00 | 4.61e-01 | 3.74e+00 | 1.81e-04 |
| “ITFG2-AS1” | 24528 | 1.20e+01 | 8.61e+02 | 2.95e+00 | 7.99e-01 | 3.69e+00 | 2.25e-04 |
| “PDZD4” | 24528 | 1.00e+00 | 6.00e+01 | -1.02e+01 | 2.77e+00 | -3.67e+00 | 2.39e-04 |
| “PHPT1” | 24528 | 1.80e+01 | 1.31e+03 | 2.39e+00 | 6.52e-01 | 3.66e+00 | 2.51e-04 |
| “TSR3” | 24528 | 2.00e+00 | 1.54e+02 | 7.10e+00 | 1.96e+00 | 3.63e+00 | 2.86e-04 |
| “CCNP” | 24528 | 2.70e+01 | 1.92e+03 | 1.91e+00 | 5.33e-01 | 3.58e+00 | 3.41e-04 |
| “ZNF692” | 24528 | 8.80e+01 | 6.21e+03 | 1.06e+00 | 2.95e-01 | 3.58e+00 | 3.50e-04 |
| “TRMT1L” | 24528 | 1.30e+01 | 9.47e+02 | 2.70e+00 | 7.67e-01 | 3.52e+00 | 4.37e-04 |
Entire population
| gene | n | sum_x | y_transpose_x | beta | standard_error | t_stat | p_value |
|---|---|---|---|---|---|---|---|
| “HMMR” | 103972 | 4.27e+02 | 2.93e+04 | -1.00e+00 | 1.30e-01 | -7.69e+00 | 1.44e-14 |
| “HOXD12” | 103972 | 6.79e+02 | 4.66e+04 | -7.80e-01 | 1.04e-01 | -7.53e+00 | 5.14e-14 |
| “PPP5C” | 103972 | 2.99e+02 | 2.04e+04 | -1.16e+00 | 1.56e-01 | -7.43e+00 | 1.11e-13 |
| “INCA1” | 103972 | 3.09e+02 | 2.12e+04 | -1.03e+00 | 1.53e-01 | -6.75e+00 | 1.50e-11 |
| “ADAP1” | 103972 | 1.45e+03 | 9.99e+04 | -4.39e-01 | 7.06e-02 | -6.22e+00 | 4.96e-10 |
| “PYGO2” | 103972 | 2.06e+02 | 1.41e+04 | -1.16e+00 | 1.88e-01 | -6.18e+00 | 6.57e-10 |
| “KMT5A” | 103972 | 2.17e+02 | 1.48e+04 | -1.11e+00 | 1.83e-01 | -6.09e+00 | 1.14e-09 |
| “CPSF3” | 103972 | 2.79e+02 | 1.91e+04 | -9.79e-01 | 1.61e-01 | -6.07e+00 | 1.28e-09 |
| “GABBR1” | 103972 | 6.56e+02 | 4.50e+04 | -6.35e-01 | 1.05e-01 | -6.03e+00 | 1.62e-09 |
| “RHEX” | 103972 | 6.49e+02 | 4.47e+04 | -6.05e-01 | 1.06e-01 | -5.71e+00 | 1.12e-08 |